Note
Go to the end to download the full example code.
Drawing a decision boundary between two interlacing moons¶
This example is a retake from the experiment in the original GEMINI paper where we want to find the true clusters in between two facing moons. To do so, the trick is to use a specific distance using the “precomputed” option which will guide the clustering algorithm to the desired solution.
Note that we use gemclus.mlp.MLPWasserstein because a linear model would not be able to find the optimal
boundary.
import numpy as np
from matplotlib import pyplot as plt
from scipy.sparse import csgraph
from sklearn import datasets, metrics
from gemclus.mlp import MLPWasserstein
Generate two interlacing moons¶
X, y = datasets.make_moons(n_samples=200, noise=0.05, random_state=2023)
Pre-compute a specific metric between samples¶
# Create an adjacency graph where edges are defined if the distance between two samples is
# less than the 5% quantile of the Euclidean distances
distances = metrics.pairwise_distances(X, metric="euclidean")
threshold = np.quantile(distances, 0.05)
adjacency = distances < threshold
# compute the all-pairs shortest path in this graph
distances = csgraph.floyd_warshall(adjacency, directed=False, unweighted=True)
# Replace np.inf with 2 times the size of the matrix
distances[np.isinf(distances)] = 2 * distances.shape[0]
Train the model¶
Note that we use the precomputed option and pass our distance to the .fit function along X.
model = MLPWasserstein(n_clusters=2, metric="precomputed", random_state=2023, learning_rate=1e-2)
y_pred = model.fit_predict(X, distances)
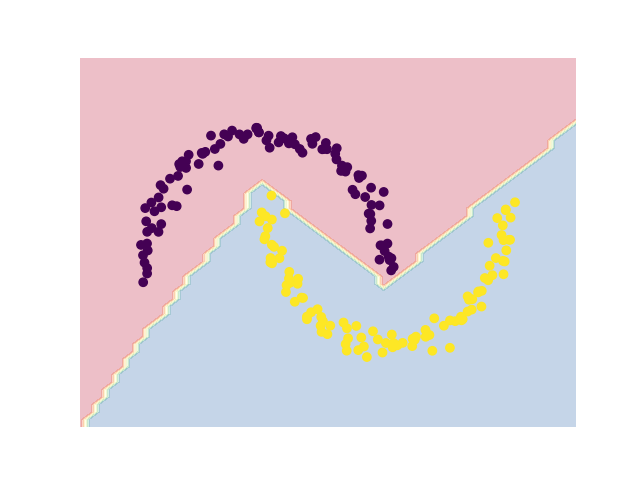
Final Clustering¶
x_vals = np.linspace(X[:, 0].min() - 0.5, X[:, 0].max() + 0.5, num=50)
y_vals = np.linspace(X[:, 1].min() - 0.5, X[:, 1].max() + 0.5, num=50)
xx, yy = np.meshgrid(x_vals, y_vals)
grid_inputs = np.c_[xx.ravel(), yy.ravel()]
zz = model.predict(grid_inputs).reshape((50, 50))
plt.contourf(xx, yy, zz, alpha=0.3, cmap=plt.cm.Spectral)
plt.scatter(X[:, 0], X[:, 1], c=y_pred)
plt.axis("off")
plt.show()
Total running time of the script: (0 minutes 7.841 seconds)