Note
Go to the end to download the full example code.
Non parametric clustering¶
This example illustrates how we can run nonparametric clustering using GEMINI. The specificity of this model is that the decision of model is not dependent on the position of the inputs, but only on the parameters associated to the input. Consequently, this model cannot be used for unseen samples as it will always return the same predictions.
import numpy as np
from matplotlib import pyplot as plt
from sklearn import metrics
from gemclus import data
from gemclus.nonparametric import CategoricalMMD
Draw samples from a GMM¶
# Generate samples on that are simple to separate
N = 100 # Number of nodes in the graph
# GMM parameters
means = np.array([[1, -1], [1, 1], [-1, -1], [-1, 1]])*2
covariances = [np.eye(2)*0.5]*4
X, y = data.draw_gmm(N, means, covariances, np.ones(4) / 4, random_state=1789)
Train the model¶
Create the Non parametric GEMINI clustering model and call the .fit method to optimise the cluster assignment of the nodes
model = CategoricalMMD(n_clusters=4, ovo=True, random_state=0, learning_rate=1e-2)
y_pred = model.fit_predict(X)
Final Clustering¶
plt.scatter(X[:, 0], X[:, 1], c=y_pred)
plt.show()
ari_score = metrics.adjusted_rand_score(y, y_pred)
gemini_score = model.score(X)
print(f"Final ARI score: {ari_score:.3f}")
print(f"GEMINI score is {gemini_score:.3f}")
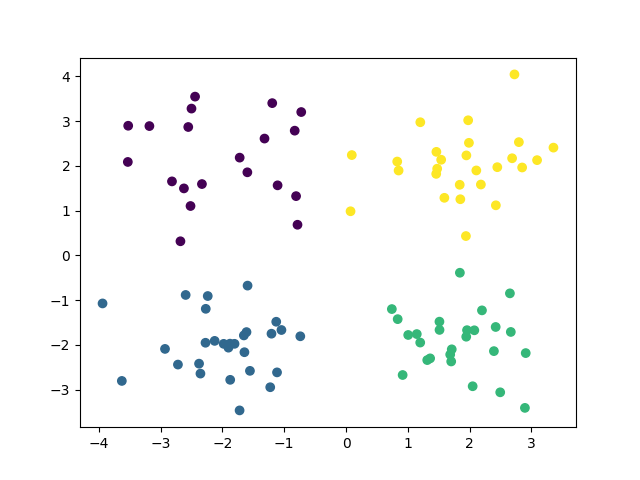
Final ARI score: 0.975
GEMINI score is 3.328
Total running time of the script: (0 minutes 0.274 seconds)